Using Electronic Health Records for Population Health Research a Review of Methods and Applications
This is an Open Admission commodity, distributed under the terms of the Creative Commons Attribution licence (http://creativecommons.org/licenses/past/4.0/), which permits unrestricted re-apply, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Electronic health records (EHRs) systems take go a crucial function of healthcare provision. Healthcare providers, health systems, and payors accept made substantial commitments to accurately capture patient information and to use EHR interfaces to amend care provision. EHRs likewise represent an important data resource as they can provide cross-sectional and longitudinal data on big "cohorts" of up to millions of individuals. Consequently, EHR data are beingness organized and leveraged to inform our understanding of disease progression, outcomes analyses, epidemiology, quality improvement, and comparative effectiveness research (CER) [Reference Chakraborty and Farooq1]. Moreover, the awarding of methods derived from statistics, computer science, data engineering, and information science has resulted in a number of high-impact findings and methodologies that take the potential to transform clinical enquiry, epidemiology, and population health sciences [Reference Goldstein, Navar, Pencina and Ioannidis2–Reference Kurant, Baron, Strazimiri, Lewandrowski, Rudolf and Digheeight].
While EHR systems represent an of import research information source, these information are highly complex and tin can exist hard to access. Typically, EHR data are stored in an enterprise information warehouse (EDW) along with a number of other information sources such equally billing and claims data, laboratory tracking systems, and scheduling data that underlie health organisation operations. These data warehouses require meaning expertise and time to navigate, and access is typically restricted to a small number of individuals to manage privacy and legal concerns associated with access to big amounts of protected wellness data (PHI). Ane way to make EHR data more than accessible and actionable for research purposes is to organize information technology into smaller relational databases, referred to as datamarts. These datamarts are typically organized nether Common Data Models (CDMs). CDMs, such equally those used by the National Patient-Centered Clinical Enquiry Network (PCORnet) and/or the Observational Medical Outcomes Partnership (OMOP), contain a set up of rules for how to turn raw EHR data into simpler data models [Reference Rosenbloom, Carroll, Warner, Matheny and Denny9]. These efforts take stimulated a significant number of retrospective analyses and innovative multicenter clinical trials [Reference Johnston, Jones and Hernandez10,11].
Objective
Due to substantial access barriers and the difficulties inherent in accessing and utilizing raw EHR data for research purposes at our establishment, we sought to develop a Knuckles University Wellness Organisation (DUHS)-specific datamart to provide well-curated and easily attainable EHR data to investigators within Duke University. In amalgam a user-facing EHR data structure, our goals were to create a data platform that
-
contains most data elements needed for clinical research studies;
-
is directly accessible by individuals conducting statistical analyses (such as the Biostatistics, Epidemiology, and Enquiry Design (BERD) cadre members or clinical scientists);
-
is accessed via a lawmaking-based system to promote reproducibility and consistency beyond studies; and
-
Utilizes a secure protected analytic workspace in which sensitive data tin can be stored and analyzed.
Later, nosotros describe the development of the Duke Clinical Research Datamart (CRDM), which contains curated and well-characterized health data that can exist accessed and analyzed using standardized methods that preserve information provenance and reproducibility. The CRDM builds off of our establishment's instance of PCORnet; as such, the CRDM is designed to support retrospective analyses of clinical data, provide reliable data with which to build study cohorts and registries, and contribute to the development of population-level wellness studies.
Materials and Methods
DUHS utilizes the Epic EHR data platform, with all wellness system data stored in an EDW, including data related to patient demographics, diagnosis and procedure codes, laboratory orders and results, medication orders and fulfillments, vital signs, encounter location information, provider notes, and other detailed clinical data. The EDW contains records for encounters at 3 Duke-affiliated hospitals and over 300 outpatient clinics, and are refreshed nightly. The Duke CRDM utilizes data within the EDW as described below.
Clarification of Data Construction
The CRDM utilizes data that have been transformed and cleaned for apply equally part of the PCORnet data network. Three hundred and xl-eight health systems from across the state participate in PCORnet, which relies on a CDM created by the Patient-Centered Outcomes Research Establish (PCORI) and licensed under Creative Commons [11], making the model freely accessible and shareable across sites. The most recent version of the PCORnet CDM (released in September 2019 [12]) defines and standardizes mapping of data elements within 22 domains. Notably, the CDM protects patient identity by assigning each patient an capricious identifier (Patient_ID) rather than a medical record number and utilizes standard terminologies to ensure consequent definitions within each domain. The CRDM utilizes the PCORnet domains shown in blue in Fig. 1. Based on Duke'south participation in the PCORnet network, the base PCORnet CDM is updated on a quarterly basis. Each domain in the CRDM is housed in a split up table (Fig. 1) in the grade of a Structured Query Language (SQL) relational database. Each tabular array contains at least ane chemical element (including Patient_ID, Encounter_ID, or Provider_ID) that allows information elements from each table to be linked together to provide specific sets of elements for a given encounter or patient.
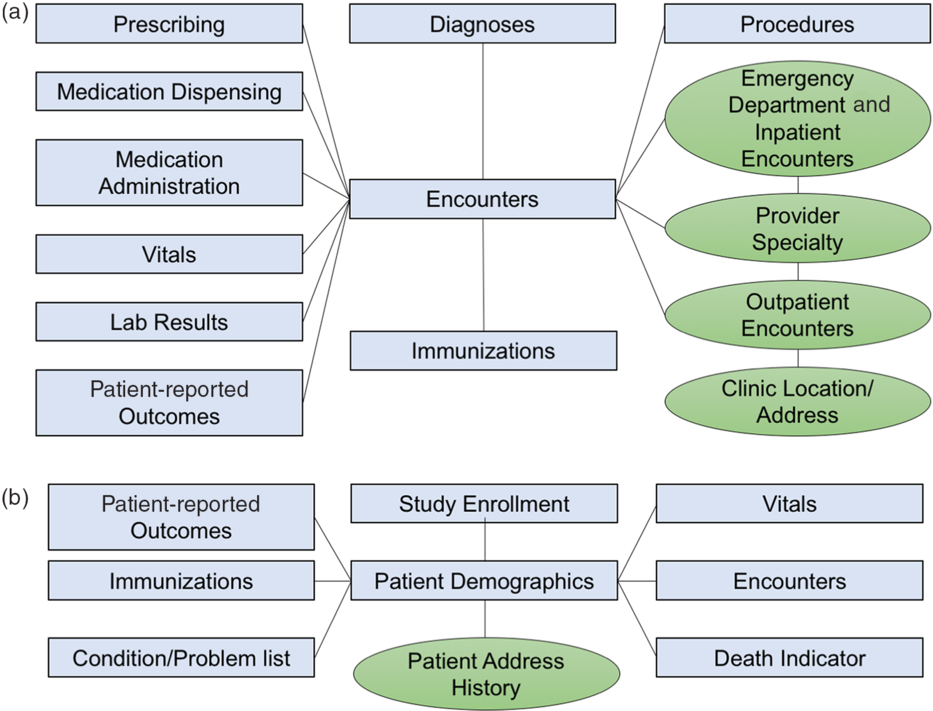
Fig. 1.Relational table structure of the Knuckles Clinical Research Datamart. (A) Encounter-linked data tables. (B) Patient-linked data tables. PCORnet-derived tables are in bluish/squares; data sidecars are shown in green/circles.
Boosted Information Elements Not Included in the PCORnet CDM
The PCORnet CDM contains nearly of the of import basic data elements for most clinical research studies; however, we found there were additional contextualizing factors that clinical researchers at our establishment desired, such as address history, dispensary location, and provider information. Therefore, we built tables for boosted data elements (referred to as "data sidecars," which are linked to and support the primary data tables). In gild to place information sidecars to be included in the CRDM, nosotros met with clinicians and investigators across disciplines and identified additional data needs through testing of different apply cases. Data sidecars that take been included in the CRDM are shown in dark-green in Fig. ane. These data elements are refreshed quarterly in conjunction with the general Knuckles PCORnet refresh so that data are comparable and linkable across both PCORnet and sidecar domains.
CRDM Employ Cases
The CRDM was designed to facilitate work within population health, comparative effectiveness enquiry/pharmacoepidemiology, predictive modeling, and the development of patient registries, which are discussed below. Instance projects are shown in Table one.
Tabular array 1. Clinical inquiry datamart utilise case examples

Population health
Population health is divers equally "the wellness outcomes of a group of individuals, including the distribution of such outcomes within a group [Reference Kindig and Stoddart13]," including health outcomes, patterns of wellness determinants, and the factors that link the ii, including policies and interventions. EHR information provide longitudinal information well-nigh large groups of individuals who receive care within a health system, making it possible to ascertain the incidence and prevalence of different diseases, disease outcomes, and the associated factors and/or exposures associated with a given disease and related outcomes. The incorporation of time-resolved addresses allows for geospatial mapping of patients to link in external data on the built surround. As an example, our group is currently using the CRDM to evaluate comorbidities, patterns of healthcare utilization, and outcomes for children with asthma who alive in Durham County, North Carolina.
Comparative effectiveness research and pharmacoepidemiology
CER using real earth data (RWD), such as data derived from EHRs, is emerging as an of import tool to evaluate the touch on of dissimilar therapeutic modalities beyond the effects that can be gleaned from randomized controlled trials (RCTs). RWD-based CER allows evaluation of a broader, more than heterogeneous group of patients than would have been included in given therapy'southward original RCT and can provide information on the touch of comorbidities, drug–drug interactions, and the acceptability and feasibility in dissimilar clinical settings. The CRDM is currently being used to evaluate the impact of opioid prescriptions on children in a postsurgical setting. Rather than randomizing a select group of patients to a specific dosing strategy later on a predefined ready of procedures, the information from the CRDM allow investigators to evaluate practice patterns in a large group of patients for an extended period of fourth dimension. Furthermore, the CRDM provides information about provider practise patterns in a nontrial setting, such that the information are reflective of normal practice patterns rather than of selected providers who are consciously collecting data, which may introduce practice bias. The CRDM is likewise being used to evaluate use of monoclonal antibodies for the treatment of severe asthma in children and adults. In the original trials, these agents were tested in study populations that were predominately white, were non large enough to evaluate the touch on of common comorbidities on efficacy, and did not provide clear guidelines to help identify patients most likely to do good from a given mAb therapeutic. The data in the CRDM provide information about the patients who are and, maybe more importantly, are not receiving mAbs, thereby providing insights into the populations that may derive the greatest benefit from these therapies.
Predictive modeling
Given the large number of individuals covered and the variety of features and outcomes captured, there is significant involvement in using EHR information for predictive modeling. Potential applications that could utilize EHR data include clinical decision support, readmission prevention, agin event avoidance, and chronic disease management. Currently, our group is using the CRDM to develop a gamble score for hospital readmissions inside thirty days of an initial hospitalization. The detailed, longitudinal data provided by the CRDM allows for rapid evaluation of risk scores using data captured past the EHR. In addition, because the patient population inclusion and exclusion criteria tin can exist readily revised, the utility of a predictive model can be evaluated across a multifariousness of patient groups.
Patient registries
Patient registries, or regularly updated lists of patients who have a particular status or meet specific clinical criteria, are powerful tools for longitudinal evaluation of wellness outcomes and utilization patterns and tin can too be used to appraise feasibility of recruiting different groups of patients for prospective trials. Once the cohorts are defined and the necessary code has been generated to assemble the dataset, the code tin can be rerun with each refresh of the datamart in lodge to call up updated patient information. We are currently building patient registries for pediatric patients with epilepsy and obesity, amid other weather.
Limitations of the CRDM
The development of the CRDM is an ongoing process that is largely defined by investigator and stakeholder identified use cases and needs. There are a few primal weaknesses of the CRDM:
-
Detailed in-hospital information: In order to simplify the data tables, the CRDM does not have time-resolved vital signs or details on transfers within the hospital.
-
Billing data: We practice not currently store whatsoever billing or claims information based on patient encounters.
-
Unstructured information: The CRDM but has structured data and does not contain any clinical notes or imaging data.
-
Real-time patient tracking: Because the CRDM is designed to be refreshed quarterly, information technology cannot be used for real-time tracking of patients. Information technology is instead suited for retrospective assessments of patient cohorts or identification of patient groups who may be eligible for recruitment into a detail clinical trial.
User Base
The CRDM's primary user base consists of statisticians and data scientists who piece of work straight with clinical researchers. Like many institutions, we have a browser-based information access layer for clinicians to query EHR data [Reference Horvath, Rusincovitch, Brinson, Shang, Evans and Ferranti14]. While these tools are useful for initial investigational work, they do non facilitate reproducible information queries. There was an expressed desire from analytic teams to be able to direct query and access EHR information. Structure Query Language (SQL) code is used to query the CRDM and database queries can exist executed using unlike programming languages, including R, SAS, and Python. The employ of SQL code to query the database ensures that the process by which data sets are created can be easily tracked, evaluated, and replicated, and SQL code tin be shared betwixt users, making information technology possible to easily recreate aspects of dissimilar data sets. Preparation modules are beingness developed by which clinical researchers with a data scientific discipline background could be trained in SQL to directly query the CRDM.
Promoting Consistency in Cohort Development and Information Retrieval
One of the challenges with EHR data is that information technology must be processed in order to be used for research. For example, there is no field within the EHR that indicates whether a patient has diabetes; instead, one has to construct a "computable phenoytpe" [Reference Spratt, Pereira and Granger15] via a combination of diagnosis codes, laboratory test results, and/or medications. While there is arguably no "all-time" phenotype definition, identification and dissemination of such phenotypes tin promote consistency beyond research studies. Nosotros are developing a lawmaking-based phenotype banking company that users tin can directly use when constructing their data sets. Sharing these and other best practices in a code-based manner ensures greater consistency beyond studies.
All registered CRDM users have access to a Gitlab page that provides a data dictionary, instructions for getting started, a list of all-time practices, and sets of SQL commands and associated accomplice definitions that can be used for practice queries. The Gitlab also serves as a central repository for each project that utilizes the CRDM. This code repository, which will grow with connected use, serves two functions: (i) to document the methods used to create analytic datasets and (ii) to provide a mechanism through which users and administrators can share known issues, all-time practices, and methodologies for creating datasets. As the code repository grows, access to this code base of operations will improve the efficiency with which users are able to gather datasets and enhance the consistency betwixt studies.
CRDM Access Environs
A major motivation behind the CRDM was to let the DUHS inquiry community the opportunity to access EHR data in a safety, consequent, and reproducible style. As such, the CRDM is housed within a server that tin only be queried while a user is logged into Duke'southward Protected Analytics Computing Environment (Step), a secure virtual network space where approved users can analyze and work with PHI and protected identifiable information (PII).
CRDM Regulatory and Admission Governance
The development of the CRDM was approved by Duke University School of Medicine'southward IRB under a database and specimens repository research summary. The IRB approved the incorporation of data held inside Duke'southward EDW as well as quarterly updates of any data held within the CRDM. The IRB allows express testing to ensure CRDM functionality, simply does not cover the use of the information for inquiry or quality improvement purposes. Any investigator who wishes to admission and use data derived from the CRDM must file a separate IRB protocol that indicates that they volition exist utilizing the CRDM and specifies which data elements volition be used and to what purposes.
Requests for access to the CRDM are initiated by filing out a CRDM Project Intake class. Users provide information nearly regulatory approvals, the data elements that will be needed for the projection, funding source, and the names of the individuals who will be querying the database. Later on ensuring that the IRB covers the stated employ and personnel, a asking is made to Duke Health Engineering science Solutions (DHTS) to grant CRDM access. The CRDM user is as well required to ship a curt narrative of the use example and to sign an understanding governing programmatic access to enterprise data resources that acknowledges all institutional policies regulating use of the data.
Dissemination and Evaluation Methods
The design of the CRDM was driven by interactions with stakeholder groups, including cores of Duke's Clinical and Translational Sciences Constitute (CTSI); representatives from the Departments of Medicine, Pediatrics, and Surgery, which together correspond the three largest clinical departments in Duke Academy School of Medicine; and an EHR datamart working grouping, which included representatives from CTSI cores, academic departments, and relevant institutes and centers from across Duke University. The working group met monthly in order to place priorities for datamart blueprint, evaluate development strategies, and discuss best practices for governance and access. CRDM blueprint was also informed by experiences garnered from airplane pilot projects spearheaded past members of the working grouping.
Members of the Duke CTSI BERD core and datamart working group also served equally the first set of CRDM exam users. The working group developed a statistical analysis plan that defined a clinical accomplice and dataset. Exam users were tasked with creating the requested dataset using the CRDM and recorded the length of time that was required, any necessary skills required for assembling the dataset, and any technical issues that were encountered during use of the CRDM. We plant that users needed to exist familiar with SQL commands, table joins, and to have some degree of familiarity with the PCORnet CDM in lodge to successfully assemble datasets.
Evaluation of the CRDM is ongoing, and the data model, governance, and access environment will exist modified in order to adjust to user and institutional needs. We are currently collecting information about the departments and divisions using the CRDM, the number of manuscripts referencing/using the CRDM, grants funded that included CRDM data, and the satisfaction of the user base. The CRDM Project Intake form currently serves every bit our primary data collection instrument, and requires that users indicate which data elements are needed for their project and any elements that are required for the projection, but that are non currently available. We will as well survey CRDM users at regular intervals to determine if they take generated inquiry products or grant applications related to the datasets that they derived from the CRDM. The datamart working group has been converted to an internal advisory board, which holds quarterly meetings to evaluate datamart use metrics (described in a higher place) and to advise on changes and upgrades to improve the usability and accessibility of the CRDM.
Give-and-take
We adult a CRDM that utilizes the existing PCORnet CDM and infrastructure in gild to create a regularly refreshed and directly attainable source of EHRs information for research and quality improvement purposes. The overarching goal of the CRDM is to serve the growing demand for EHRs-derived datasets to exist used in population health studies, comparative effectiveness research, and quality comeback, and to provide an efficient and reproducible mechanism for amalgam EHR datasets. Furthermore, by expanding EHR information access, we hope to increment the pool of investigators, statisticians, analysts, and trainees working with these data in society to answer questions related to health, patient well-being, and healthcare delivery.
The CRDM utilizes resource that have been developed equally part of the Clinical and Translational Science Awards (CTSA) program and could exist replicated by other institutions with CTSAs. Bookish medical centers that participate in PCORnet have the clinical inquiry data network infrastructure that is a critical component of the CRDM, every bit the data take already been cleaned and curated and therefore attach to a widely used data model. In order to use these data to create a CRDM-like structure, each center would demand to work with their information engineering science departments to identify a platform where these data could exist stored that would exist both HIPAA-compliant and straight accessible to approved investigators and enquiry staff. DUHS has developed the Footstep, which is a secure virtual computing environment for this purpose. Similar environments are becoming available at academic medical centers to facilitate the apply of patient data while providing data security. The data sidecars have been designed to provide site-specific contextual details while harmonizing with the PCORnet CDM, such that the sidecars direct link to and can be refreshed with the rest of the PCORnet instance. Other centers could define their own sidecars and would also be able to replicate the sidecars from the Knuckles CRDM by sourcing data from similar elements in their own data warehouses. In addition, the CRDM user base primarily consists of CTSA cores, including the BERD core and informatics core. These cores are also present at other CTSAs and can be leveraged to build data resources. The CRDM model can therefore be replicated at most large academic medical centers.
Future work with the CRDM volition focus on training of an expanded user base, dissemination of best practices, and evaluation of user metrics. We are currently developing presentations and online learning modules to provide data about the CRDM that volition help potential users to place the best data resource for their research questions and quality improvement projects. We are also evaluating how all-time to train BERD cadre members and other statistics and analytics trained groups, including clinician researchers, in the use of the CRDM, including SQL coding and best practices for assembling patient cohorts and analytic datasets. Every bit described above, we are beginning to track the use of the datamart, including any enquiry products and grant applications supported by its use.
Expanding the pool of investigators utilizing EHR information to answer research questions is 1 of the principal goals of the development of the CRDM. While providers spend a pregnant corporeality of time inputting data into EHR systems, many accept non had the opportunity to use these data to respond clinical enquiry questions. Moreover, the assay of EHR data tin be complex, and it is important to understand how data capture mechanisms should influence the estimation of results. In society to help familiarize investigators with the use of EHR data for research purposes, we are working with Knuckles's CTSI and the Clinical Inquiry Training Program to develop a set up of didactic courses that will serve as an introduction to research uses of EHR data. Such training sessions could potentially be shared across CTSA sites that are using similar data models. In addition, nosotros are actively working to pair investigators from different clinical specialties with informaticians, biostatisticians, and researchers who accept previously used EHR data in order to address specialty-specific topics. These pairings will serve to raise the skill sets of all investigators past serving as a conduit for sharing cognition most the apply of health data and clinical questions that tin exist investigated using this new data resource.
Decision
The CRDM was developed with the input of multiple stakeholder groups from beyond Duke University School of Medicine and leverages existing information resources, including the DUHS electronic data warehouse, the PCORnet CDM, and the data that have been cleaned and standardized for incorporation into the CRDM for use in the Duke PCORnet case. Moreover, the CRDM is available to support CTSA cores including Bioinformatics and BERD, among others. Given that these resource are available at other Clinical and Translational Sciences Accolade sites, we believe that the evolution of the CRDM could be readily replicated at other academic medical centers.
Acknowledgments
The authors thank the Duke Electronic Health Records Datamart Working Group, Knuckles Health Engineering science Solutions, Duke Children'south Health and Discovery Initiative, and the Knuckles Clinical and Translational Sciences Special Populations Core for their support and guidance in the development of the CRDM.
Funding was provided past Duke Wellness, Knuckles Academy School of Medicine, the Duke Department of Pediatrics via the Translating Duke Health Children'south Health and Discovery Initiative, the NCATS CTSA grant (UL1TR002553), and the NHLBI (5R21HL145415).
Disclosures
The authors have no conflicts of involvement to declare.
References
Chakraborty, P , Farooq, F. A robust framework for accelerated consequence-driven risk factor identification from EHR. In: Proceedings of the 25th ACM SIGKDD International Conference on Cognition Discovery & Data Mining – KDD '19. New York: ACM Press; 2019. 1800–1808.CrossRefGoogle Scholar
Goldstein, BA , Navar, AM , Pencina, MJ , Ioannidis, JPA. Opportunities and challenges in developing take chances prediction models with electronic health records data: a systematic review. Journal of the American Medical Informatics Association 2017; 24(ane): 198–208.CrossRefGoogle ScholarPubMed
Xiao, C , Choi, East , Sun, J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. Journal of the American Medical Informatics Association 2018; 25(x): 1419–1428.CrossRefGoogle ScholarPubMed
Hemingway, H , Asselbergs, FW , Danesh, J , et al. Big information from electronic health records for early and late translational cardiovascular inquiry: challenges and potential. European Heart Journal 2018; 39(sixteen): 1481–1495.CrossRefGoogle ScholarPubMed
Schinasi, LH , Auchincloss, AH , Forrest, CB , Diez Roux, AV. Using electronic health record data for environmental and place based population health enquiry: a systematic review. Annals of Epidemiology 2018; 28(7): 493–502.CrossRefGoogle ScholarPubMed
Jensen, Pb , Jensen, LJ , Brunak, Southward. Mining electronic wellness records: towards better inquiry applications and clinical intendance. Nature Reviews Genetics 2012; 13(half-dozen): 395–405.CrossRefGoogle ScholarPubMed
Shortreed, SM , Cook, AJ , Coley, RY , Bobb, JF , Nelson, JC. Challenges and opportunities for using big health intendance information to advance medical science and public wellness. American Journal of Epidemiology 2019; 188(5): 851–861.CrossRefGoogle ScholarPubMed
Kurant, DE , Baron, JM , Strazimiri, 1000 , Lewandrowski, KB , Rudolf, JW , Dighe, AS. Cosmos and use of an electronic health record reporting database to improve a laboratory test utilization program. Applied Clinical Computer science 2018; 9(3): 519–527.Google ScholarPubMed
Rosenbloom, ST , Carroll, RJ , Warner, JL , Matheny, ME , Denny, JC. Representing knowledge consistently across wellness systems. Yearbook of Medical Information science 2017; 26(i): 139–147.Google ScholarPubMed
Johnston, A , Jones, WS , Hernandez, AF. The ADAPTABLE trial and aspirin dosing in secondary prevention for patients with coronary artery disease. Current Cardiology Reports 2016; eighteen(eight): 81.CrossRefGoogle ScholarPubMed
Horvath, MM , Rusincovitch, SA , Brinson, S , Shang, HC , Evans, S , Ferranti, JM. Modular design, awarding architecture, and usage of a cocky-service model for enterprise data delivery: the Knuckles Enterprise Data Unified Content Explorer (DEDUCE). Journal of Biomedical Informatics 2014; 52: 231–242.CrossRefGoogle Scholar
Spratt, SE , Pereira, One thousand , Granger, BB , et al. Assessing electronic health tape phenotypes against gold-standard diagnostic criteria for diabetes mellitus. Journal of the American Medical Informatics Association 2017; 24(e1): e121–e128.CrossRefGoogle ScholarPubMed
Source: https://www.cambridge.org/core/journals/journal-of-clinical-and-translational-science/article/development-of-an-electronic-health-records-datamart-to-support-clinical-and-population-health-research/5422C3AD02D5785C810E54CD9A7F76D9
Post a Comment for "Using Electronic Health Records for Population Health Research a Review of Methods and Applications"